Technology
We're building towards foundation models for genomics and chemical perturbation of cells. We take a dynamical systems view towards providing a mechanistic understanding of how drugs act at the cellular level.
To guide drug discovery and design, the Syntensor platform allows the user to simulate a variety of preclinical assays. We have prioritized predicting potency and toxicity endpoints, as collectively, these two areas account for 60-70% of clinical trial failures. The predictions of these assays are contextualized with genome-wide predictions of changes in gene expression and pathway activation.
The Syntensor platform predicts and explains the outcomes of preclinical assays.
Preclinical assays have low information content and often yield conflicting results, leading to high uncertainty about the mechanism of action of a drug beyond its primary target.
If scientists were able to understand all of the pathways that were perturbed by a drug, they could design a more informative assay strategy and anticipate issues with efficacy and toxicity before progressing a candidate to clinical trials.
To meet this challenge, the Syntensor Platform leverages predictive models trained on mutlti-omics and experimental data to simulate cellular assays at scale and contextualize assay outcomes with predicted genome-wide changes in gene expression levels for novel compounds across cell lines. These changes are further summarized at the pathway level so that users can understand how perturbing the primary target of the drug leads to changes in the state of the cell. Users can visualize these predictions in the Pathway Explorer to generate testable hypotheses about how their drugs are perturbing cellular signaling pathways downstream of binding the primary target of the drug.
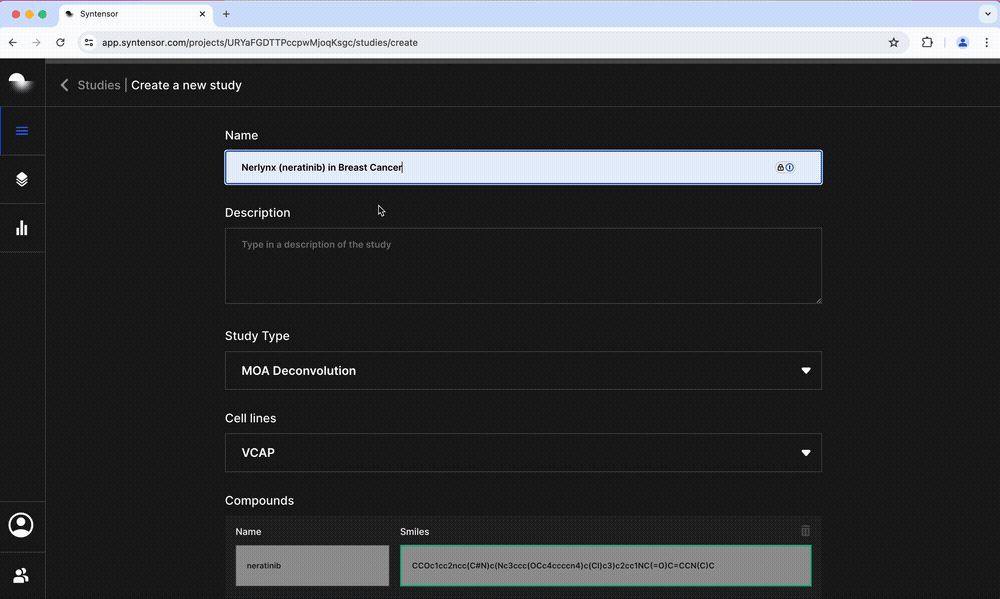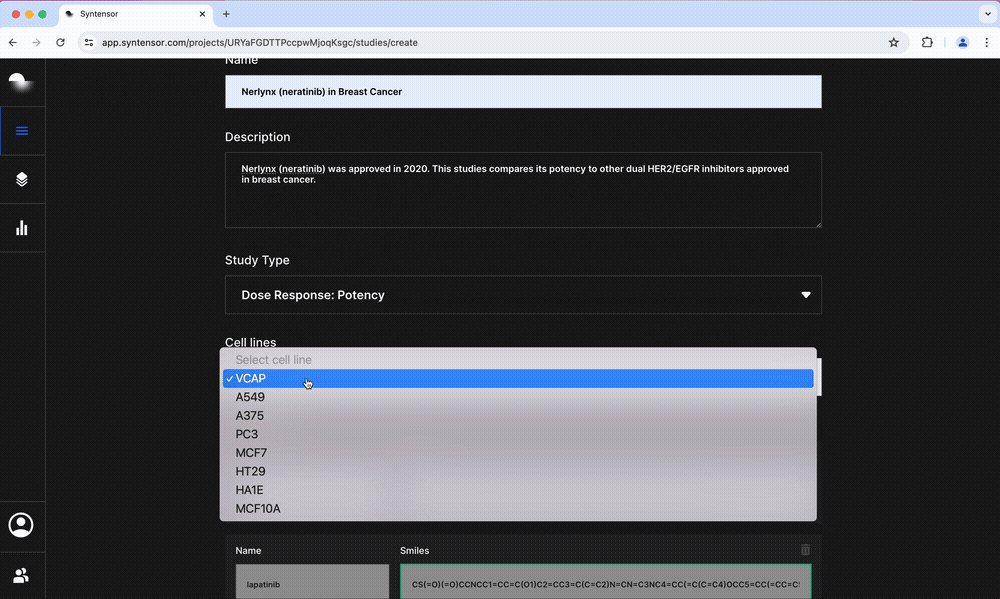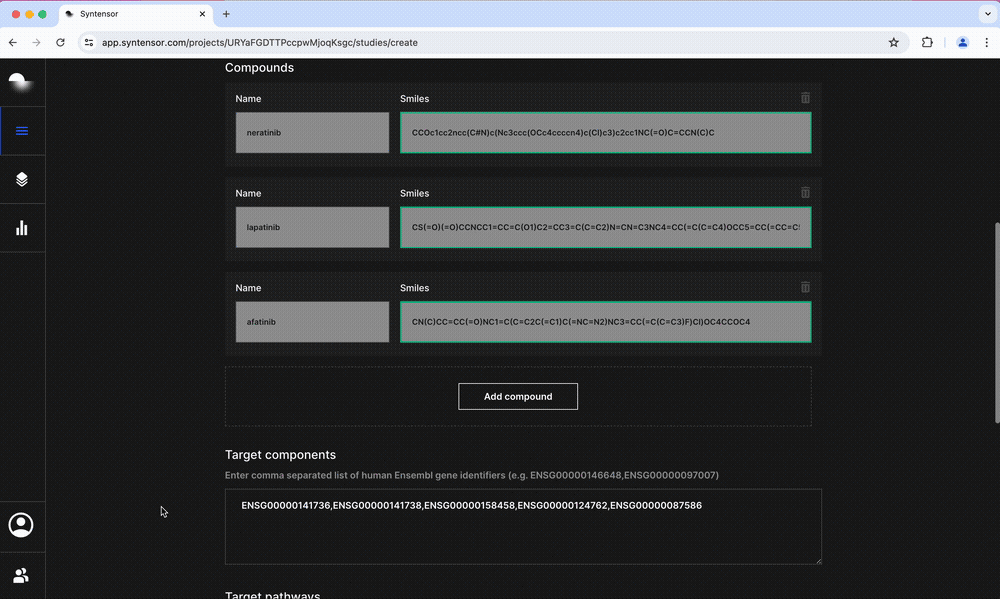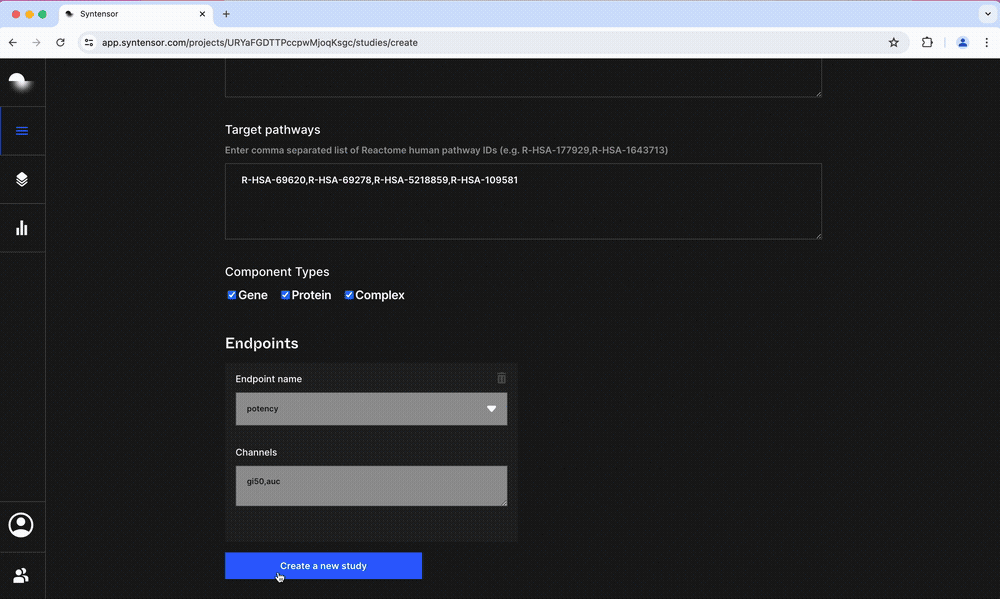
- Mechanism of Action: Generates a ranked list of potential MoA targets by taking into account all perturbed pathways
- Potency: Predicts growth inhibition and sensitivity in cancer cell lines and ties these to changes in apoptotic pathways
- Toxicity: Characterizes the potential for hepatotoxicity by predicting the possibility of drug-induced liver injury and results for 14 assay endpoints including CYP450 inhibition and mitochondrial toxicity